We very recently surveyed our CORE members to ask what was most important to them and we received wide-ranging feedback. The CORE dashboard provides a range of tools for our data providers and their repository managers and users. Much of the feedback we received was regarding providing additional or enhanced tools for managing repository content via the dashboard. For example, metadata validation and enrichment tools were regarded as highly important.
Interestingly however, what was most important was making repository content machine-readable. This is closely linked to identifying funding information and rights-retention strategies. Ensuring content is machine-readable allows for the extraction of far richer information from full-text documents than that available in the metadata alone. In the U.S., the recent OSPT memo on ‘Ensuring Free, Immediate, and Equitable Access to Federally Funded Research‘ includes machine-readability as a required component of the archiving and deposition of federally funded research.
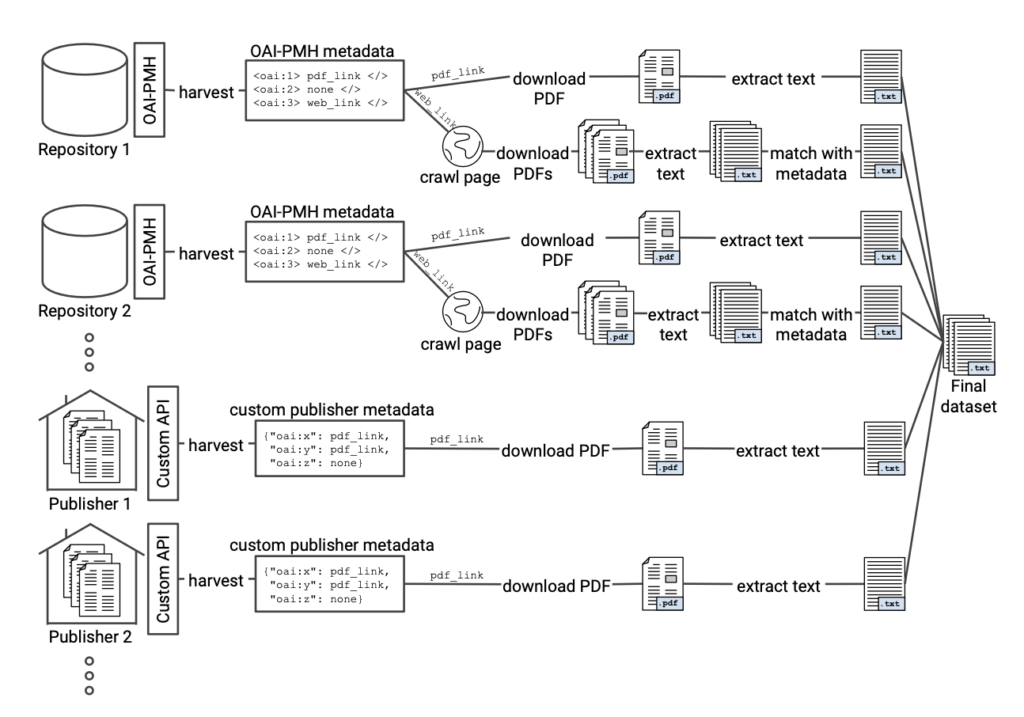
The figure below maps how CORE currently aggregates content from the global network of almost 11,000 repositories. As part of this process the raw text is extracted from the PDF documents and is then available with the PDF documents as part of the regularly released CORE datasets and is also available via the CORE API
Example illustration of the data collection process. The figure depicts the typical minimum
steps which are necessary to produce a dataset for TDM of scientific literature.
This raw text is however unstructured and can be noisy and difficult to work with at scale. We’re really happy therefore to announce the work we have been doing to provide structured representations of full-text scientific documents using the open source tool, GROBID.
GROBID (GeneRation Of BIbliographic Data) is a machine learning library for extracting, parsing and re-structuring raw documents such as PDF into structured XML/TEI encoded documents with a particular focus on technical and scientific publications. Developed by Patrice Lopez, originally in 2008, GROBID was made open source in 2011 and has continued to be supported and developed until now. It provides a wide range of tools for parsing PDF documents;
- Header extraction and parsing from article in PDF format. The extraction here covers the usual bibliographical information (e.g. title, abstract, authors, affiliations, keywords, etc.).
- References extraction and parsing from articles in PDF format, around .87 F1-score against on an independent PubMed Central set of 1943 PDF containing 90,125 references, and around .90 on a similar bioRxiv set of 2000 PDF (using the Deep Learning citation model). All the usual publication metadata are covered (including DOI, PMID, etc.).
- Citation contexts recognition and resolution of the full bibliographical references of the article. The accuracy of citation contexts resolution is between .76 and .91 F1-score depending on the evaluation collection (this corresponds to both the correct identification of the citation callout and its correct association with a full bibliographical reference).
- Full text extraction and structuring from PDF articles, including a model for the overall document segmentation and models for the structuring of the text body (paragraph, section titles, reference and footnote callouts, figures, tables, etc.).
- PDF coordinates for extracted information, allowing to create “augmented” interactive PDF based on bounding boxes of the identified structures.
- Parsing of references in isolation (above .90 F1-score at instance-level, .95 F1-score at field level, using the Deep Learning model).
- Parsing of names (e.g. person title, forenames, middle name, etc.), in particular author names in header, and author names in references (two distinct models).
- Parsing of affiliation and address blocks.
- Parsing of dates, ISO normalized day, month, year.
- Consolidation/resolution of the extracted bibliographical references using the biblio-glutton service or the CrossRef REST API. In both cases, DOI/PMID resolution performance is higher than 0.95 F1-score from PDF extraction.
- Extraction and parsing of patent and non-patent references in patent publications.
GROBID is highly efficient at processing documents and here at CORE we have a lot of documents. Like a *lot*. The CORE archive currently consists of approximately 34 million full-text scientific articles. We are using the Big Data Cluster here at KMi, which is a Kubernetes cluster, and so far we’ve processed just over 20 million documents. These are already available via the CORE API with a specific endpoint for retrieving the structured tei.xml file. Details can de found in the API documentation.
Once we have completed the GROBID-ification (!) of the current CORE collection, a new dataset will be made available. Additionally, GROBID will be added to the ingestion pipeline for all new documents aggregated by CORE.
Patrice Lopez, the creator of GROBID said;
“It’s really exciting to see GROBID being used at this scale on the whole CORE collection. Having this structured representation of scientific documents facilitates further text mining processes and can be the source of additional enrichments – so CORE and GROBID together offer a clear added value here over just having PDF documents.”
Petr Knoth, founder and team lead at CORE added;
“Access to large scale corpora of scientific articles in a structured format allows for some really interesting and novel use-cases. Adding GROBID as part of the ingestion process at CORE means that, in the future, all PDF documents will have this structured format available. This additional information means we can deliver solutions for our data providers and members that capitalises on the data extracted by GROBID such as funding information, author affiliations and citations. Further, researchers, scientists, developers and other stakeholders can now use this enhanced data in their own applications.”
For more information on GROBID you can visit the GitHub repository for the project.